Overview
PatchMAN (Patch-Motif AligNments) is a novel peptide docking approach which uses structural motifs to map the receptor surface with backbone scaffolds extracted from protein structures.
Protocol Outline
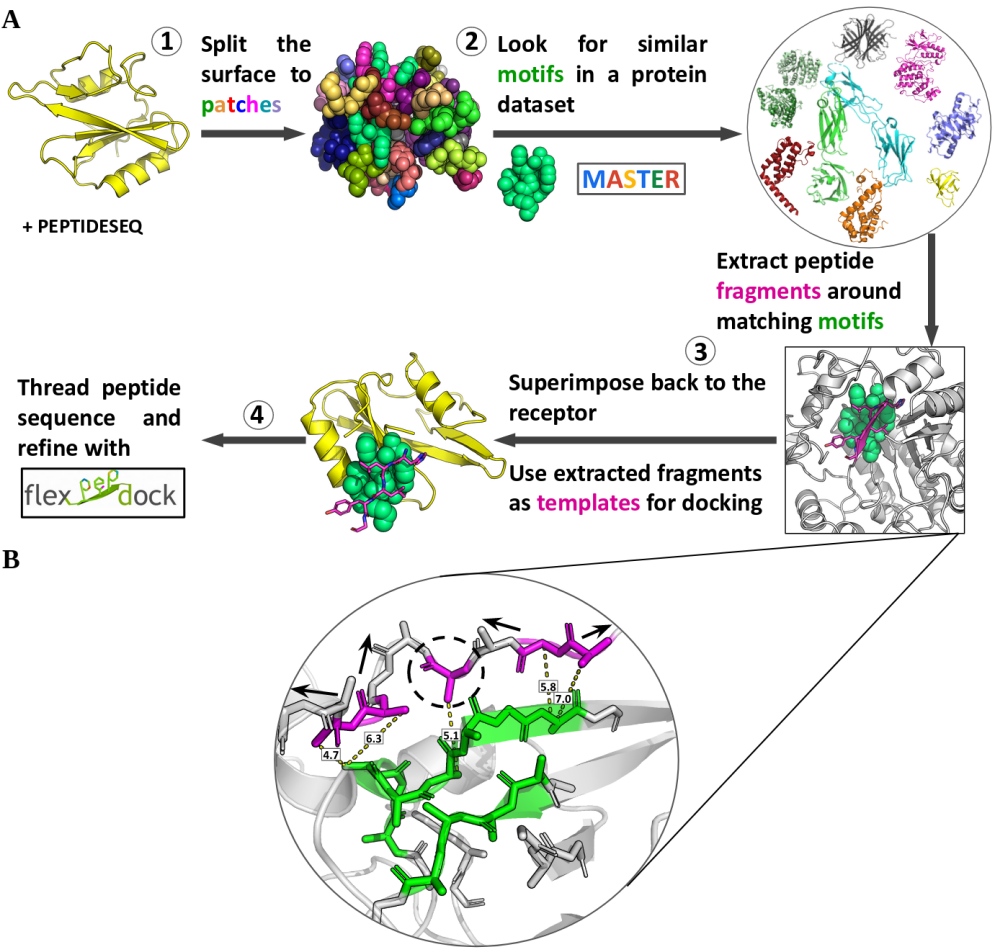
Scheme of the The PatchMan protocol.
(A) Flowchart. The input is a receptor PDB file and a peptide sequence.
(1) Definition of surface motifs on the receptor: The protein surface is defined based on solvent accessibility,
and then split into small structural surface patches.
(2) Identification of structural matches in protein structures: Matches are detected using MASTER search against
a non-redundant dataset of protein structures;
(3) Generation of the peptide-protein complex structure: the peptide fragment is determined (see (B))
and superimposed onto the receptor. Then the peptide sequence is threaded onto the identified complementing fragment;
(4) Refinement and scoring: the initial structures are refined using the Rosetta FlexPepDock refinement protocol,
and top-scoring models are selected as final predictions.
(B) Extracting peptide fragments. Neighboring residues (magenta) around the matching motif (green) are defined
with Cβ distance within 8Å of the motif. Consecutive backbone stretches are then elongated in both directions
to the desired peptide length. Arrows indicate stretches that can be elongated. Single residue
(indicated with dashed circle) will not be elongated.